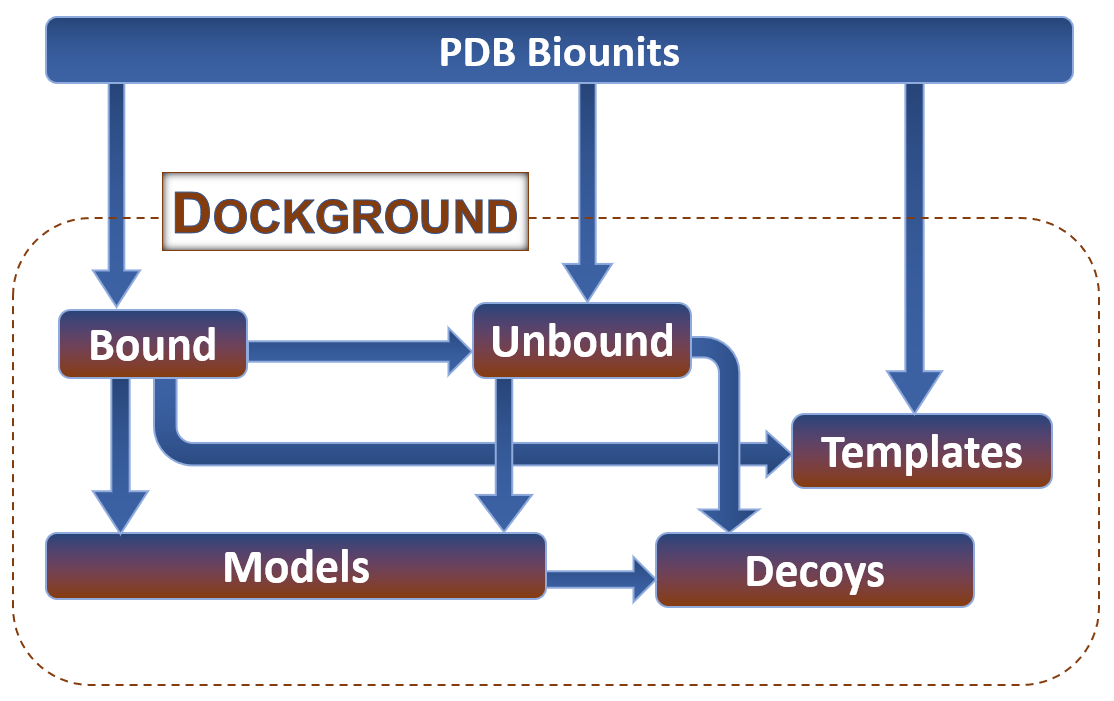
Datasets
Bound
Unbound
Models
Decoys
Templates
Assessment
Rotamers
Other Resources
Downloads
Docking Benchmark Set 1 15.6MB, 92 complexes
Docking Benchmark Set 2 30.4MB, 242 complexes
Docking Benchmark Set 3 31.0MB, 233 complexes
Docking Benchmark Set 4 50.7MB, 396 complexes
Docking Benchmark Set 1 92.9MB, 3,205 complexes
Docking Benchmark Set 1 287.8MB, 63x6 complexes
Docking Benchmark Set 2 59.2MB, 165x6 complexes
Docking Benchmark Set Q1 55.4MB, 171x5 complexes
Docking Benchmark Set Q2 325.1MB, 963x5 complexes
X-ray Unbound Set 1 519.8MB, 101x61 complexes
X-ray Unbound Set 2 855.3MB, 100x396 complexes
Model Set 1 2.4GB, 100x165x6 complexes
Full Structures 1.0 374.0MB, 5,050 complexes
Interfaces 1.0 206.5MB, 7,107 complexes
Full Structures 1.1 390.6MB, 4,950 complexes
Interfaces 1.1 194.8MB, 5,936 complexes
Full Structures 2.0 1.1GB, 12,470 complexes
Interfaces 2.0 404.5MB, 12,431 complexes
Repository Status
2/26/2026
Docking Benchmarks
| Set | Number of complexes |
|---|---|
| X-ray Unbound Set 1 | 92 |
| X-ray Unbound Set 2 | 242 |
| X-ray Unbound Set 3 | 233 |
| X-ray Unbound Set 4 | 396 |
| Simulated Unbound Set 1 | 3205 |
| Models Set 1 | 63 x 6 |
| Models Set 2 | 165 x 6 |
| Models Set Q1 | 171 x 5 |
| Models Set Q2 | 963 x 5 |
Docking Templates
| Set | Number of complexes |
|---|---|
| Full Structures 1.0 | 5050 |
| Interfaces 1.0 | 7107 |
| Full Structures 1.1 | 4950 |
| Interfaces 1.1 | 5936 |
| Full Structures 2.0 | 12470 |
| Interfaces 2.0 | 12431 |
Docking Decoys
| Set | Number of complexes |
|---|---|
| X-ray Unbound Set 1 | 101 x 61 |
| X-ray Unbound Set 2 | 100 x 396 |
| Model Set 1 | 100 x 165 x 6 |
References
DOCKGROUND should be cited as:
Collins, K.W., Copeland, M.M., Kotthoff, I., Singh, A., Kundrotas, P.J., and Vakser, I.A., 2022, DOCKGROUND resource for protein recognition studies, Prot. Sci., 31: e4481.Other DOCKGROUND publications:
- Douguet D, Chen H, Tovchigrechko A, Vakser IA Dockground resource for studying protein-protein interfaces. Bioinformatics 2006, 22:2612-2618
- Gao Y, Douguet D, Tovchigrechko A, Vakser IA Dockground system of databases for protein recognition studies: Unbound structures for docking. Proteins 2007, 69:845-851
- Liu S, Gao Y, Vakser IA Dockground protein-protein docking decoy set. Bioinformatics 2008, 24:2634-2635
- Anischenko I, Kundrotas PJ, Tuzikov AV, Vakser IA Protein models: The Grand Challenge of protein docking. Proteins 2014, 82:278-287
- Anischenko I, Kundrotas PJ, Tuzikov AV, Vakser IA Protein models docking benchmark 2. Proteins 2015, 83:891-897
- Anischenko I, Kundrotas PJ, Tuzikov AV, Vakser IA Structural templates for comparative protein docking. Proteins 2015, 83:1563-1570
- Kirys T, Ruvinsky AM, Singla D, Tuzikov AV, Kundrotas PJ, Vakser IA Simulated unbound structures for benchmarking of protein docking in the Dockground resource. BMC Bioinformatics 2015, 16:243
- Kundrotas PJ, Anishchenko I, Dauzhenka T, Kotthoff I, Mnevets D, Copeland MM, Vakser IA Dockground: A comprehensive data resource for modeling of protein complexes. Protein Sci 2018, 27:172-181
- Singh, A, Dauzhenka, T, Kundrotas, PJ, Sternberg, MJE, Vakser, IA. Application of docking methodologies to modeled proteins. Proteins. 2020; 88: 1180–1188.
- Kundrotas, P.J., Kotthoff, I., Choi, S.W., Copeland, M.M., Vakser, I.A., 2020, DOCKGROUND tool for development and benchmarking of protein docking procedures, Methods in Mol. Biol., 2165:289-300.
- Kotthoff, I., Kundrotas, P.J., Vakser, I.A., 2022, DOCKGROUND membrane protein-protein set, PLoS ONE, 17: e0267531.
- Kotthoff, I., Kundrotas, P.J., and Vakser, I.A., 2022, DOCKGROUND scoring benchmarks for protein docking, Proteins, 90:1259-1266.
- Collins, K.W., Copeland, M.M., Brysbaert, G., Wodak, S.J., Bonvin, A.M.J.J., Kundrotas, P.J., Vakser, I.A., Lensink, M.F., 2024, CAPRI-Q: The CAPRI resource for assessment of modeling protein interactions, J. Mol. Biol., doi:10.1016/j.jmb.2024.168540.